Home
Biophysical Motivation
Installation
Quick Start Jupyter Notebook
Quick Start command line
Examples
Publications
Quickstart Guide for the Command Line
-
Download and install Anaconda Distribution for Python
-
Open terminal and type:
pip install molclustpy
-
Create a working directory - /user/Desktop/molclustpy
-
Download the pair of bngl and python files (For example, EGFR_model.bngl and EGFR_model.py) and place them inside the working directory
Downloadable python input files
Downloadable bngl files
- Nephrin_Nck_NWASP_high_concentration.bngl
- Tutorial_Nephrin_Nck_NWASP.bngl
- EGFR_model.bngl
- Using the terminal, navigate to the working directory and type
python EGFR_model.py
Due to the system configuration, in some cases, you may need to type
python3 EGFR_model.py
-
The script should sequentially import the package, run multiple stochastic trials, analyze and visualize the data
-
Note: when you run python from the terminal, the figures will appear one after the other, that is, the second figure is displayed only after you close the first figure and so on. If you run python from an IDE like Spyder, all the figures are sequentially displayed, just like the jupyter notebook.
-
Change the simulation parameters and visualize the results
Detailed description of code-blocks
This notebook provides a step by step guide on how to change simulation parameters and analyze the results.
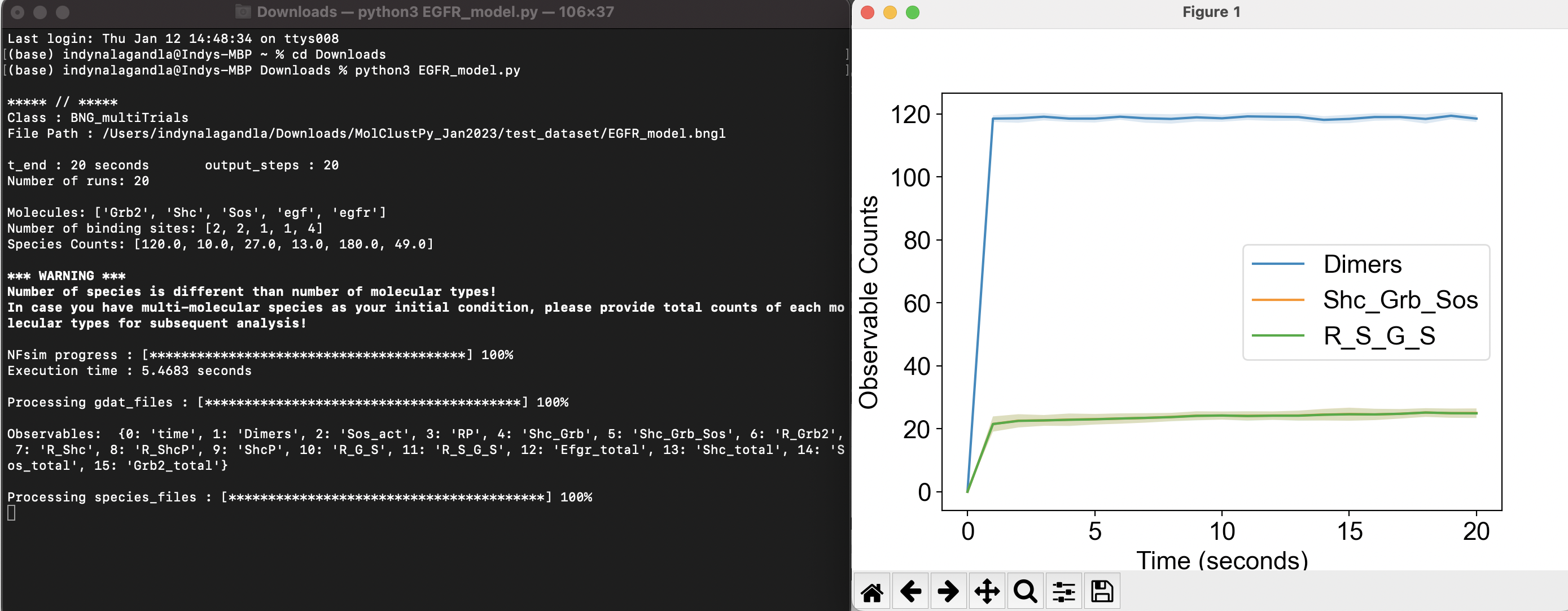
Terminal Output and Results
The terminal will ouput this:
The outputed results will be found in the same location as the bngl file:
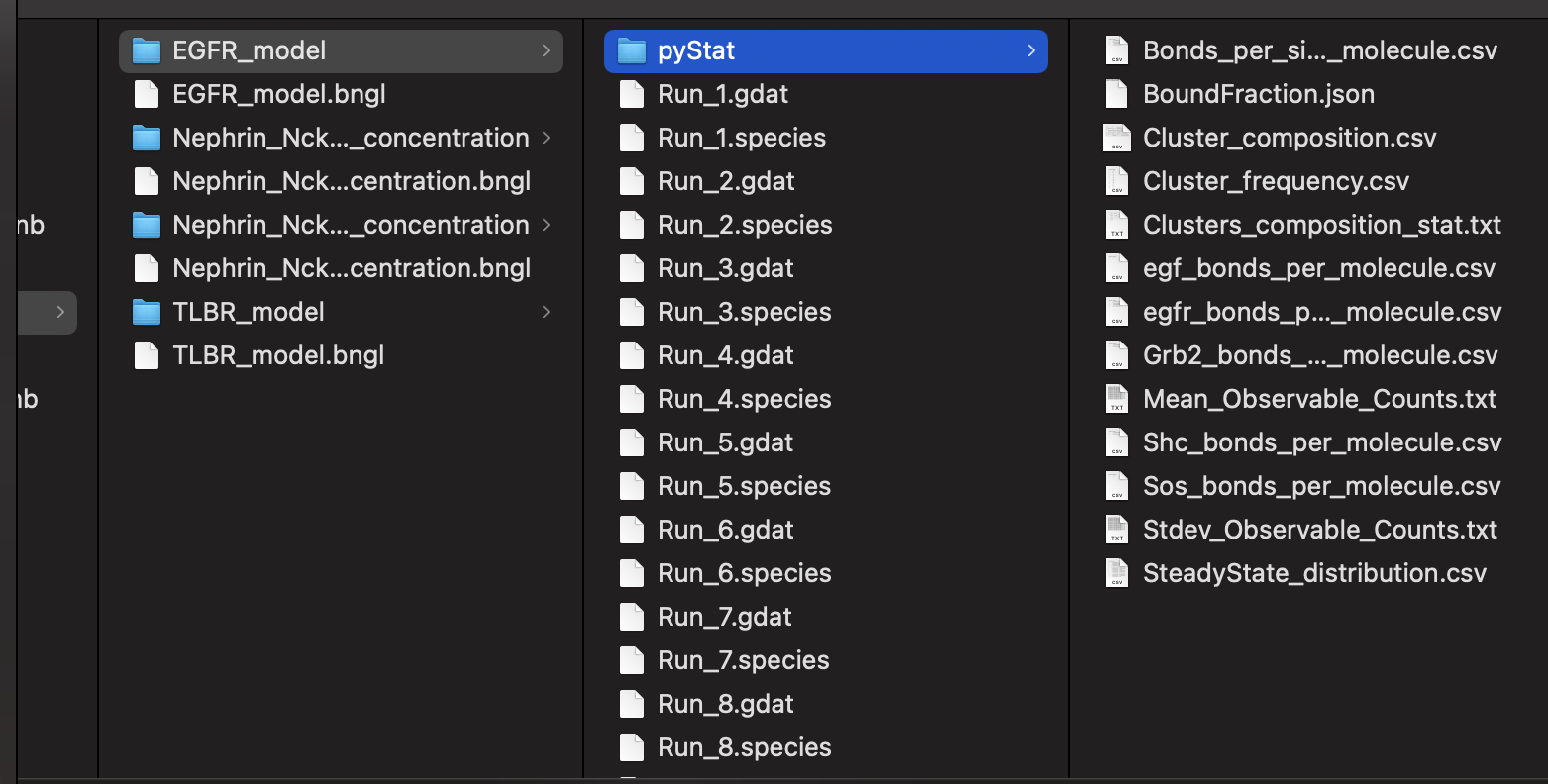
Simulation output will be stored in a folder called MyModel if the model name is MyModel.bngl. There are two types of output:
- gdat files (Run_1.gdat, Run_2.gdat, … , Run_N.gdat) containing the timeseries of observables
- species files (Run_1.species, Run_2.species, … , Run_N.species) containing the molecular species (clusters)
Note: If the folder already contains results and number of current trials is less than existing ones, then existing results will be deleted. For higher number of trials, existing trajectories will be overwritten.
Locating Package Through Command Line
If you need to find the location of the package
- open terminal
- open python and type
import molclustpy
print(molclustpy.__file__)
This will output a path to the package that will look something like this
/usr/local/lib/python3.10/site-packages/molclustpy/__init__.py
Common Error Identification
- We sometimes encounter an error involving Bionetgen (likely due to the version control issues)
AttributeError: module 'bionetgen' has no attribute 'bngmodel'
- In such cases, the following resolves the issue
pip uninstall bionetgen
pip install bionetgen