Home
Biophysical Motivation
Installation
Quick Start Jupyter Notebook
Quick Start command line
Examples
Publications
Higher concentrations of Nephrin, Nck and NWASP produce large clusters, an indication of phase transition.
It is recommended that you run Jupyter notebook - it has all the text below. This page is given just for convenience of those who don’t have an opportunity to run notebook
# Package import
from NFsim_data_analyzer import *
from DataViz_NFsim import *
from MultiRun_BNG import *
# bngl file (BioNetGen model)
bng_file = './test_dataset/Nephrin_Nck_NWASP_high_concentration.bngl'
# Initialization of the Simulation Object
simObj = BNG_multiTrials(bng_file, t_end=0.02, steps=20, numRuns=20)
print(simObj)
simObj.runTrials(delSim=False)
print()
***** // *****
Class : BNG_multiTrials
File Path : ./test_dataset/Nephrin_Nck_NWASP_high_concentration.bngl
t_end : 0.02 seconds output_steps : 20
Number of runs: 20
Molecules: ['NWASP', 'Nck', 'Nephrin']
Number of binding sites: [6, 4, 3]
Species Counts: [300.0, 900.0, 450.0]
NFsim progress : [****************************************] 100%
Execution time : 4 mins 58 secs
# analyze data across multiple trials
outpath = simObj.getOutPath()
molecules, numSite, counts, _ = simObj.getMolecules()
nfsObj = NFSim_output_analyzer(outpath)
print(nfsObj)
nfsObj.process_gdatfiles()
nfsObj.process_speciesfiles(molecules, counts, numSite)
***** // *****
Class : NFSim_output_analyzer
System : Nephrin_Nck_NWASP_high_concentration
Total Trajectories : 20
Processing gdat_files : [****************************************] 100%
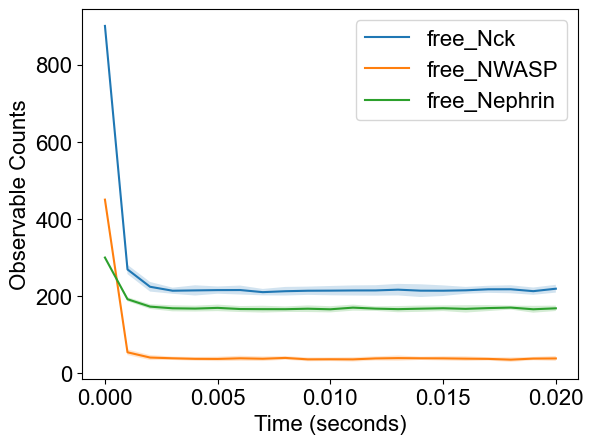
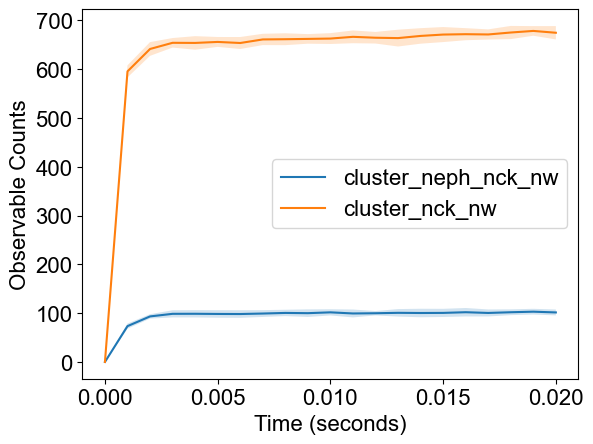
Observables: {0: 'time', 1: 'tot_Nck', 2: 'free_Nck', 3: 'tot_NWASP', 4: 'free_NWASP', 5: 'tot_Nephrin', 6: 'free_Nephrin', 7: 'fully_bound_Nephrin', 8: 'fully_bound_Nck', 9: 'fully_bound_NWASP', 10: 'cluster_neph_nck_nw', 11: 'cluster_nck_nw'}
Processing species_files : [****************************************] 100%
# Visualization
# indexList for observables
plotTimeCourse(outpath, obsList=[2,4,6])
plotTimeCourse(outpath, obsList=[10,11])
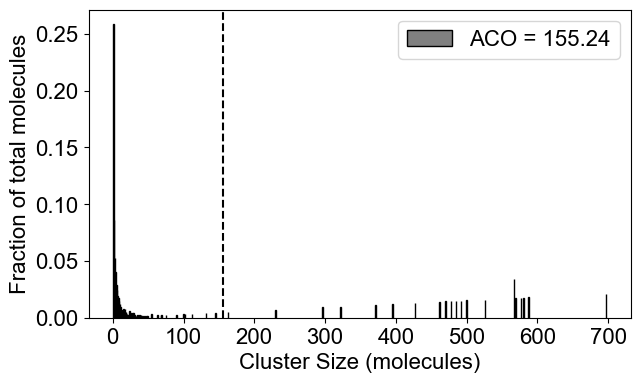
# 2A: Cluster size distribution (ACO: Average Cluster Occupancy)
plotClusterDist(outpath)
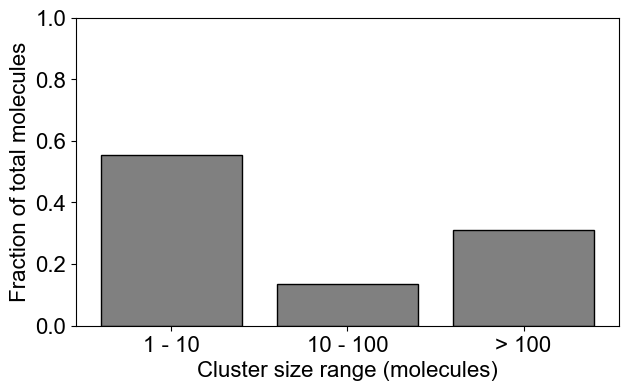
# Binned distribution by providing cluster size ranges
plotClusterDist(outpath, sizeRange=[1,10,100])
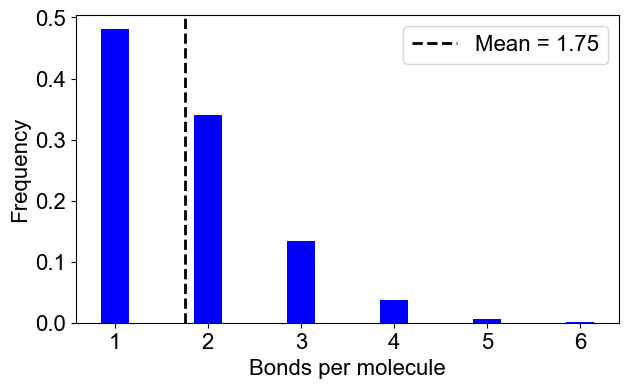
# 2B: Number of bonds per molecule
plotBondsPerMolecule(outpath)
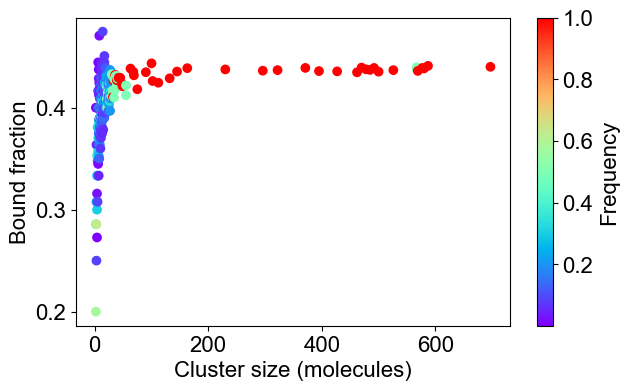
# 2C: Bound fraction distribution
plotBoundFraction(outpath)
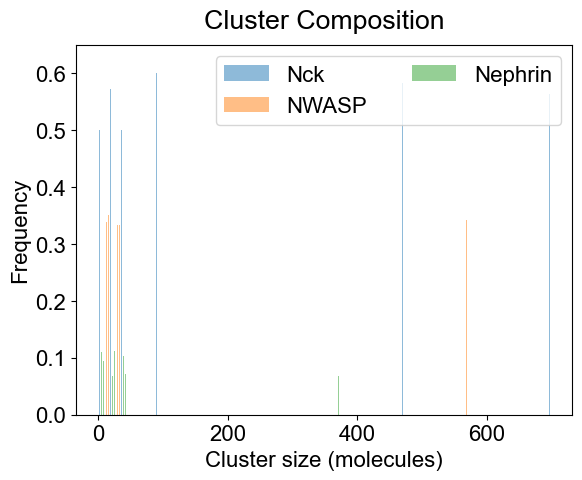
# 3A. Average composition of indivual clusters.
# Default is all the clusters present in the system. As before, adjust width and transparency (alpha) for visual clarity.
plotClusterComposition(outpath, specialClusters=[], width=0.15, alpha=0.5)
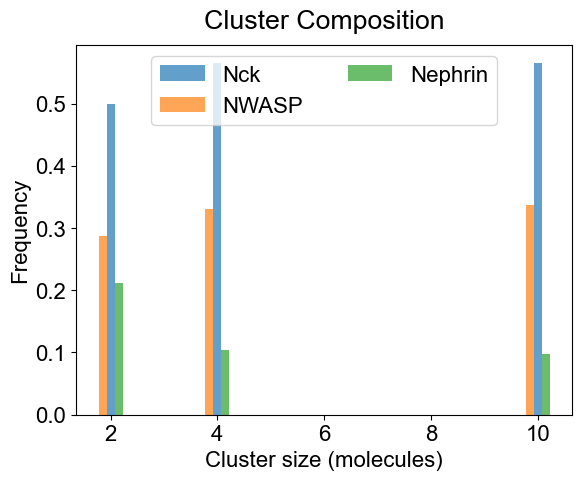
# You can look at the composition of a set of clusters (specialClusters) also
plotClusterComposition(outpath, specialClusters=[2, 4, 10], width=0.15, alpha=0.7)
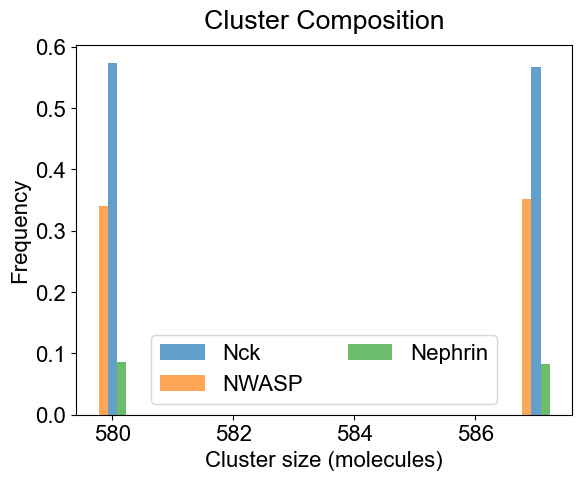
plotClusterComposition(outpath, specialClusters=[580,587], width=0.15, alpha=0.7)
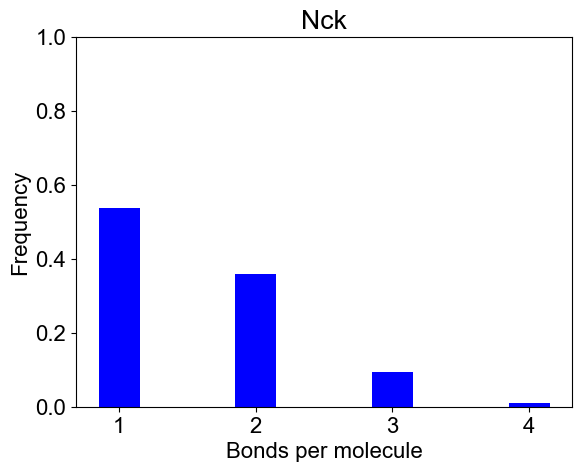
# 3B. Bondcount distribution of each molecular type
# plotBondCounts(outpath, molecules=molecules) # Reading molecules from previous block
# You may provide a subset of molecules also
plotBondCounts(outpath, molecules=['Nck'])